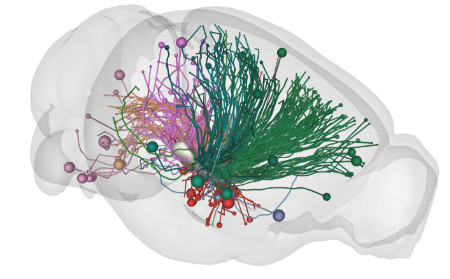
Visualisation: BrainRender
This Python-based package handles visualization of three dimensional neuroanatomical from publicly available datasets (e.g. Allen Brain atlas) and from user generated experimental data. The goal of brainrender is to faciliate the exploration and communication of neuroanatomical data by providing a user-friendly platform to create 3D renderings.
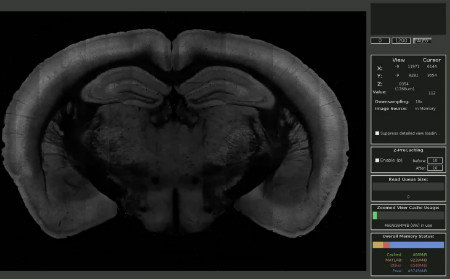
Visualisation: MaSIV
This MATLAB-based package loads downsampled image stacks but presents the user with full-res data as they zoom in. MaSIV is stable but can only display a single channel at once. Extendable via plugins.
Visualisation: Lasagna
This Python-based software is currently under development. It provides three linked orthogonal 2-D views for fast visualisation of downsampled image stacks. Allows overlays of multiple brains, multiple channels, traced neurites, or soma locations. Includes viewer for Allen Atlas. Extendable via plugins.
Links
- Napari - Fast visualisation of big image data in Python
- Fiji - You can use virtual stacks or the BigDataViewer to navigate large datasets
- Icy - Fiji alternative
- ClearVolume - Fiji plugin for making pretty transparent volumes.
- spimagine - Volume renderer
- Fluorender - 3D volume renderer aimed at neuroscientists. Probably can’t handle large datasets.
- VAA3D - Multi-dimensional data viewer for big datasets. Can be hard to set up but is powerful.
- MIB - General purpose MATLAB-based viewer for quick visualisations. No big data options.
- Knossos - Big data image annotator
- Orbit - Image analysis tool
- Free Imaris Viewer - Imaris now have a stripped down version that is free