Registering widefield functional imaging data to the Allen Atlas
Is it possible to register the brain surface as seen in widefield in vivo imaging to the Allen Atlas?
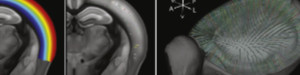
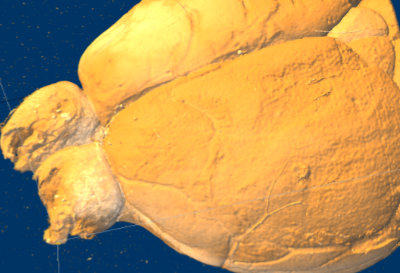 Displaying downsampled serial section data in Fiji’s volume viewer shows that at least larger blood vessels are visible on the surface rendering. Ivana Orsolic and Petr Znamenskiy decided to use the vessels to register widefield functional imaging data to serial section stacks and so to the Allen Atlas.
Displaying downsampled serial section data in Fiji’s volume viewer shows that at least larger blood vessels are visible on the surface rendering. Ivana Orsolic and Petr Znamenskiy decided to use the vessels to register widefield functional imaging data to serial section stacks and so to the Allen Atlas.
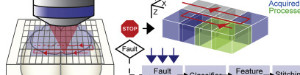
The brain was sliced and imaged, then the downsampled volume was registered to the Allen Atlas as usual. The 3D image stack is now in the same space as the Allen Atlas. Using Petr’s recoverVasculature function found in StitchIt the blood vessel patterns (now also in the Atlas space) are extracted from the downsampled serial section data. An affine transformation based on the vessels brings the in vivo data into the same space as the sample and hence into the same space as the Atlas.

References
This work was inspired by the Allen Institute informatics white paper on serial 2p processing.